UK wild Arabidopsis
Allaby Research Group and Holub Research Group

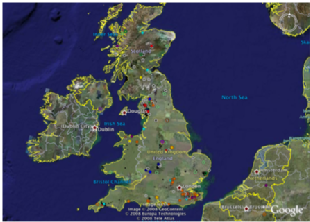
While we can use archaeogenetics to examine evidence of local adaption through time in species such as barley, for modogenetic approaches it makes a lot of sense to use the model system Arabidopsis thaliana. Why Arabidopsis and not some 'natural' plant or crop? Well, it is a naturally occurring weed species, a possible human commensal in the British Isles, for which there is a formidable amount of data available all wrapped up in a bite size genome. It is possible to do ecological genomics with this organism in a way which is simply not possible for any other species of plant or animal right now: it is a natural trail blazer. In conjunction with the Holub group we are interested in the wild UK Arabidopsis population, for which we have genome wide SNP information. Interestingly, the UK population is highly structured, indicating long term occupancy of certain localities. Other areas have 'hybrid' populations, possibly representing 'suture' zones of expansion of different populations. We wish to study how natural variation in these populations correlates with selection pressures. How does the genome as a whole adapt to mural environment or a disease rich environment? Are cohorts of genes involved, or even entire regulatory systems? To answer these sorts of questions we will be using association mapping approaches for instance, as well as using the TreeMos tool to study the 'lumpiness' of gene flow across the genome between populations in order to look for signs of selection.
