Functional analysis of regulatory genes involved in Arabidopsis leaf senescence
Leaf senescence is a programmed event responding to a wide range of external and internal signals. It requires de novo gene expression and protein synthesis and is controlled in a tightly regulated manner. Many different genes show enhanced expression during senescence and elucidation of the roles of these genes is throwing light on the mechanisms that occur during the senescence process. Elucidation of the genes that control senescence has been complicated by the complex combination of signalling pathways that appear to be involved in senescence. Cross talk exists between senescence and stress or pathogen responses and also hormonal and nutrient signals are implicated in the control of senescence.
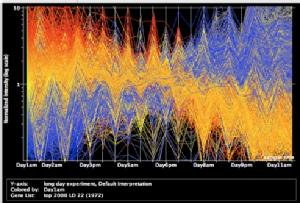
Using micorarray analysis using RNA from leaves sampled at many timepoints during development, we have identified several thousand genes that are upregulated or down regulated during leaf senescence in Arabidopsis. This graph shows the top 2000 most differentially expressed genes over the timecourse. Leaves were harvested at two different times of day, the diurnal expression of many of these is evident.
Functional analysis of senescence enhanced regulatory factors
Within the upregulated genes, we have identified a large number of putative regulatory genes that may function to control gene expression during senescence. We are using insertion mutants and RNAi lines to investigate the effects of knocking out the function of individual genes. Also over expression of genes in transgenic Arabidopsis is being used to assess the function of each gene. Data from the Systems Biology project will identify further genes for functional analysis.

The effect of each knockout or overexpression mutant is being examined using CATMA microarrays. These arrays carry sequence tags for over 30 000 Arabidopsis genes. Altered expression patterns for groups of genes indicate the signalling pathway perturbed by the mutation.
Mutant analysis
Some of the knock out mutants have an effect on senescence related parameters.
For example, insertion in two different stress related genes that show senescence enhanced expression results in premature senescence in response to stress. IM38 and IM100 are two different mutants with insertions in the same senescence enhanced heat shock transcription factor. Plants subjected to drought stress show premature senescence

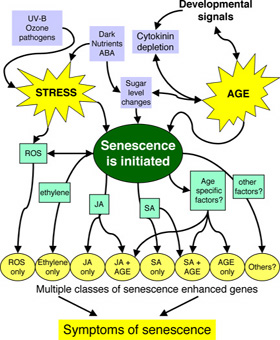
Using the results of the insertion mutant analysis we have a preliminary model for the pathways that regulate the senescence process. This will be supplemented by transcriptional network information from the Systems Biology programme. We aim to identify the key branch points in the signalling pathways that can be manipulated to alter senescence.
Vicky Buchanan-Wollaston
Emily Breeze
Philip Law
Adam Talbot
Christine Hicks