Genetic Markers
Genetic markers are used to identify different features in DNA sequence that can be used to differentiate between individuals in a population, or to classify individuals between different varieties or cultivars within a species. The different features in the sequence can be used to identify if that particular region was inherited from the female or male parent. By using this information we can build up a more complete picture of each individuals genotype at each marker. This allows us to track the inheritance of different regions of the genome.
Types of molecular genetic markers
There are many, but some of the commonly used ones are below:
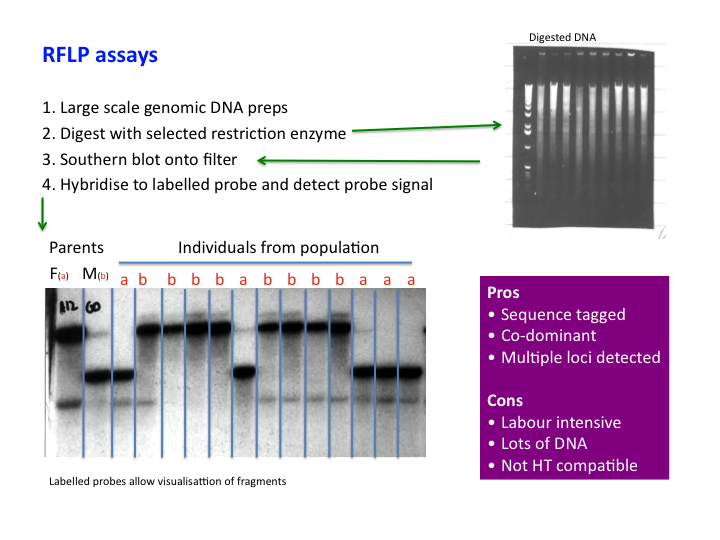
RFLP – Restriction Fragment Length Polymorphism
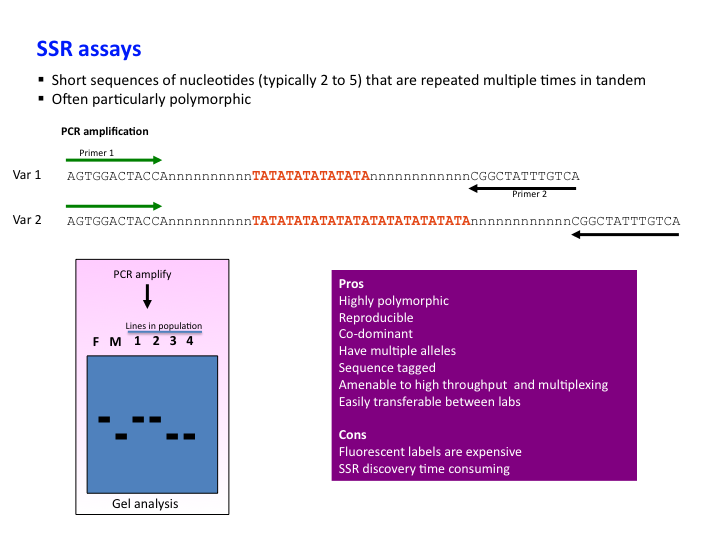
SSR – Simple Sequence Repeat, or Microsatellite
AFLP – Amplified Fragment length Polymorphism
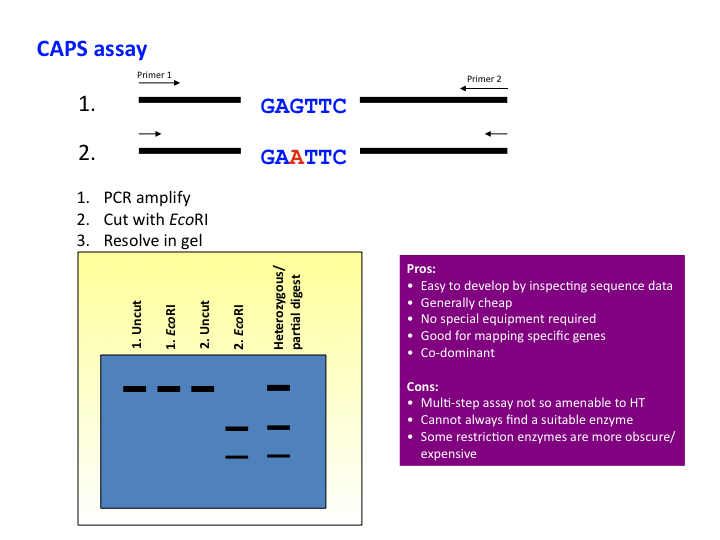
CAPS – Cleaved Amplified Polymorphic Sequence
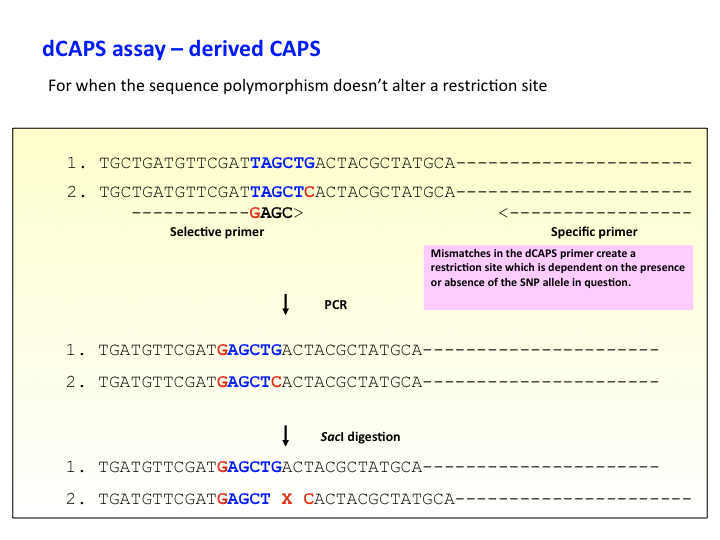
dCAPS – derived Cleaved Amplified Polymorphic Sequence
STS – Sequence Tagged Site
RAPD – Ramdomly Amplified Polymorphic DNA
SNP – Single Nucleotide Polymorphism - technology to screen thousands in one go
Also Phenotypic markers
Genetic markers: RFLP
Genetic markers: SSR assay
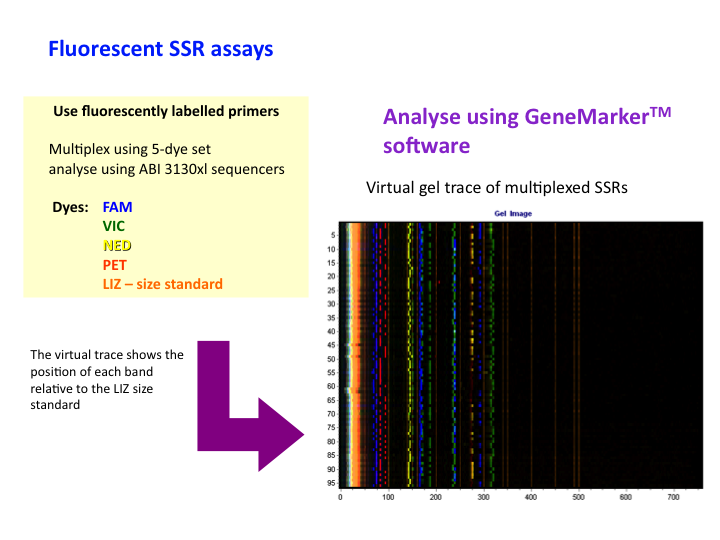
Genetic markers: Fluorescent SSRs
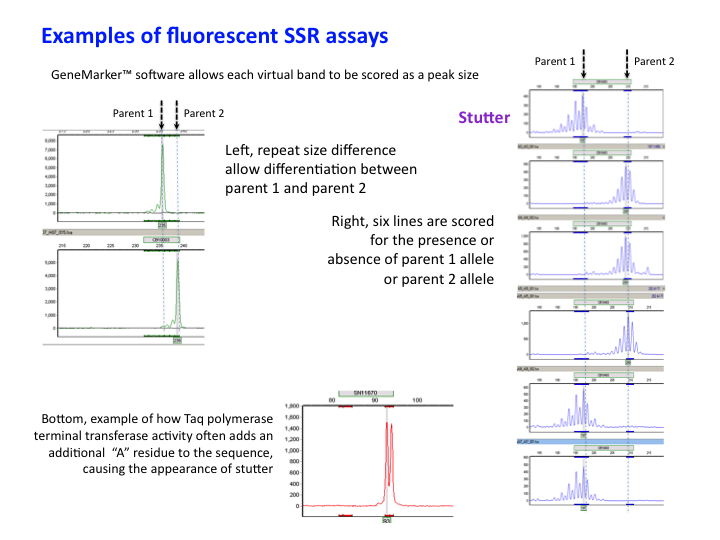
Example Labelled SSR assay
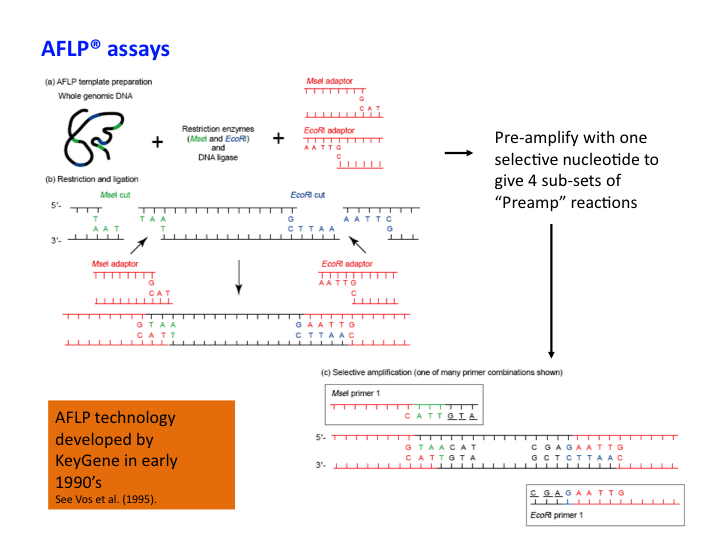
Genetic markers: AFLP
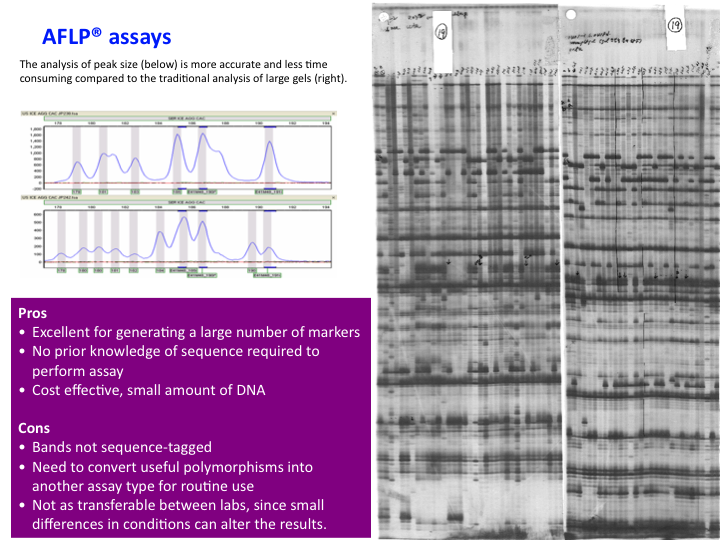
AFLP Results
Genetic markers: CAPS assay
Genetic markers: derived CAPS
 |
|
Genetic markers: SNPs
The most common subunits of DNA are the four deoxyribonuclotides (nucleotides) containing the bases: adenine (A), cytosine (C), guanine (G) and thymine (T). A single Nucleotide Polymorphism (SNP) describes a single base difference between two DNA sequences. For example, a C/T substitution in the DNA of plant 2 compared to the same region of DNA in plant 1
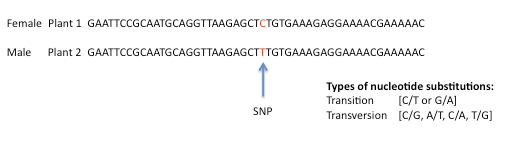
Using DNA sequence data from the parents of a population, SNPs can be identified and used to screen their progeny – the individuals in the population.
For example 8 members of a population derived from a cross between two parents above.
C/T SNP scored in 8 lines

Plants 4 and 7 have inherited the polymorphism from the male donor. If the SNP was in the coding sequence of a gene this may alter the phenotype.
Since the nucleotide is the smallest unit in DNA, SNPs represent the most abundant genetic marker type. New sequencing technologies enable thousands of SNPs to be discovered. These new markers have revolutionised the resolution attainable for genetic maps, and enhanced the precision of association analysis. SNPs used in genetic maps can be used to align sequence data to their respective chromosome positions. This is particularly useful in the absence of a fully assembled genome, and is essential during genome assembly and validation.
