Georgina Charlton
Background
BSc Hons Chemistry 2011-2014 University of Warwick
MSc AS:MIT 2015-2016 University of Warwick
PhD MAS CDT 2016-2020 University of Warwick
Postdoc 2021-Present UCL
PhD Project
Life sciences project with Dr Alex Jones on " Proteomic Analysis of Drought & Oxidative Stress in Arabidopsis thaliana & Zea mays" sponsored by Syngenta.
Plant stress occurs when a plant is growing in non-ideal conditions, which put an increased demand on its ability to function. There are many different types of stress such as salt, temperature, drought, pests, herbicides and so on. These stresses will affect metabolism, root development, reproduction or growth.
The Effect of Drought on Maize Yield
Maize (Zea mays) is a cereal crop in the grass family which produces kernels of seed which are used for food for humans and livestock as well as biofuels. Maize plants usually develop 20-21 leaves during their lifetime, flower (silk) 65 days after sprouting and the cobs are fully mature after 120 days (Figure 1). These time scales and number of leaves can vary depending on variety, environment and the time of year the crop is planted. Global maize production between 2008 and 2010 was 750 million metric tons making maize the world’s third-largest crop for food production, after wheat and rice, and any effect on its growth cycle leading to loss of yield can be devastating to the food industry especially in developing countries.

Figure 1: Here's how the maize plants have progressed from planting to maturity.
The tandem mass tag (TMT) consists of an amine-reactive group, which reacts with the N terminus of peptides, a mass normaliser region, and a reporter ion, which fragments during MS2 giving diagnostic peaks which can be used for quantification (Figure 1). The amine-reactive group also reacts with basic amino acids, such as lysine residue, as well as the N terminus of tryptic peptides. The TMT tags add up to the same overall molecular weight but have heavy isotopes (C13 and N15) in different ratios within the reporter and mass normaliser regions of the molecule, so will have different mass reporter ions. Proteins are digested and the peptides are tagged with TMT, each sample reacted with a different TMT molecule can then be combined and analysed using LC-MS/MS, the ratios of the reporter ions in each spectrum give quantitative information on the proteins in the samples. Because the tags have the same overall mass and hydrophobicity they cannot be distinguished until MS2.
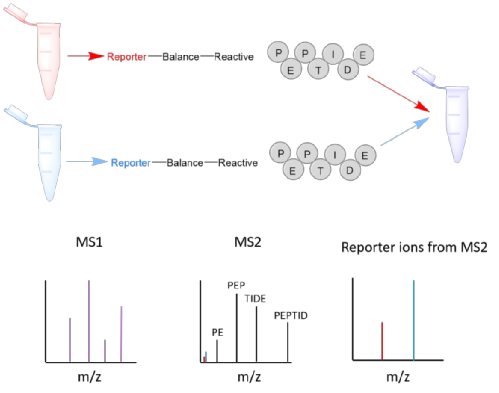
Figure 2: TMT tags and how they work.
Post-translational modifications (PTMs) are biochemical modifications which occur on amino acids after the protein has been synthesised. PTMs change the function of proteins by adding a functional group, proteolytic cleavage of regulatory subunits, or degradation of proteins. PTMs play an important role in cell function. Protein phosphorylation is one of the most important PTMs, which causes loss, gain or change in function of proteins. Serine, threonine and tyrosine amino acid residues are the most common sites for phosphorylation. Protein phosphorylation is reversible and plays an important role in cell signalling. Sequential phosphorylation of proteins plays an important part in the signalling cascade such as the canonical MAPK signalling module. However, due to the modification’s low levels in the proteome, phosphorylation modifications have proved difficult to analyse directly and some form of enrichment is needed. The most common method for phosphopeptide enrichment involves metal ion chelation, which uses the metal ions’ positive charge to interact with the negatively charged phosphate group, allowing phosphate groups to be enriched from complex mixtures (Figure 3).
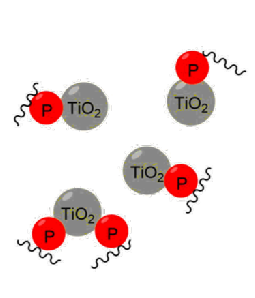
Figure 3: Phosphopeptide enrichment.
During my work, the effect of drought on maize yield was studied using TMT and mass spectrometry, the effect of drought on phosphorylation modifications was also studied. I was able to identify 1114 proteins in maize leaves, of which 271 proteins were identified which changed significantly during drought stress. I was able to identify drought response proteins including abscisic acid (ABA) response proteins and dehydrins which significantly changed due to drought treatment. In the phosphorylation experiment, I was able to identify 589 phosphopeptides of which 61 phosphopeptides were identified in the leaves which significantly changed in the drought treated samples. The phosphopeptides which significantly increased in abundance mapped to proteins associated with drought and stress response
The Effect of Methyl Viologen on Arabidopsis thaliana
Arabidopsis thaliana was established as the model plant due to its short lifespan, small size, high seed production and high transgenic efficiency and was the first plant to be fully genome sequenced in 2000. Arabidopsis encodes 25,498 genes, and other plant genomes were completed soon after. The availability of these databases allowed proteome analysis to become easier by enabling high throughput assignment of peptide sequences to tandem mass spectra. Mass spectrometry-based proteomics allows qualitative and quantitative analysis of high numbers of proteins from organisms. Mass spectrometry also allows information to be gained on post-translational modifications (PTMs), protein-protein interactions, and protein function.
Methyl viologen (MV) is the active ingredient in Paraquat, a wide spectrum herbicide used for weed control in agriculture which kills plants by causing oxidative stress. MV reactions take place in the chloroplasts of plant cells and cause a decrease in the amount of chlorophyll a and b in cells, decreasing leaf pigment and photosynthetic activity (Figure 4).
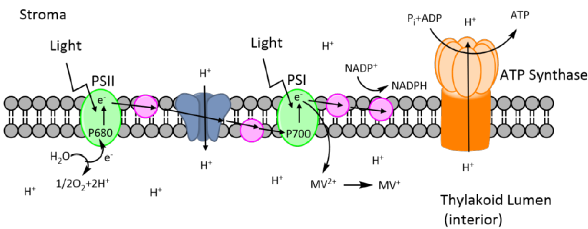
Figure 4: Methyl viologens mechanism of attack at photosystem II during photosynthesis.
Using TMT for quantitative analysis I was able to identify 1342 proteins in the Arabidopsis soluble fraction of which 397 significantly changed during treatment. The proteins which significantly increase in abundance included oxidative stress proteins. I was able to identify 879 proteins in the Arabidopsis membrane fraction of which 578 proteins significantly changed in abundance during MV treatment. The proteins which significantly increase in abundance included oxidative stress proteins.
Phosphopeptide analysis of MV treated Arabidopsis was also performed. 1082 phosphopeptides were identified in the Arabidopsis soluble fraction of which 84 significantly changed during the treatment. The phosphopeptides which increase in abundance mapped to proteins associated with stress, photosynthesis and MAPK activity. 761 phosphopeptides were identified in the Arabidopsis membrane fraction of which 42 significantly changed due to the treatment. The phosphopeptides which significantly increased in abundance mapped to proteins associated with stress, photosynthesis and kinase activity.
Development of a Method for Tagging & Enrichment of Carbonyl Modifications
Stress in plants can be caused by a variety of factors and create changes to proteins in the form of post-transitional modifications (PTMs ). Reactive oxygen species (ROS) mediate cell signalling pathways but under stress conditions, their quantities become too high and they cause oxidative damage to DNA, lipids and proteins. Oxidative damage causes irreversible damage to proteins, leading to loss or change in function. Carbonylation modifications are used as key biomarkers of oxidative stress in animals and plants due to their relative chemical stability. However, due to their low abundance, carbonyl modifications have been difficult to analyse. Therefore, a suitable tagging and enrichment process is needed to fully analyse carbonyl modifications. Enrichment can be performed at the protein level or the peptide level (Figure 5). Protein level enrichment will give information on the proteins which are modified but because most of the peptides are unmodified, it will not give information on the site which is modified. Peptide level enrichment will give information on the site which is modified but will increase the false discovery rate because a 1 peptide tolerance needs to be used when analysing data.
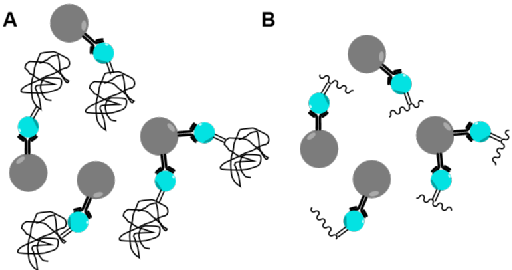
Figure 5: (A) Enrichment at the protein level. (B) Enrichment at the peptide level.
I used two different chemistries to perform carbonyl enrichment experiments, hydrazines form hydrazone bonds ( N-N=C) with carbonyl groups alkoxyamine reagents form oxime bonds (O-N=C) (Figure 6). The hydrazone bond is unstable under acidic conditions, which means an extra reduction step needs to be performed to stabilise the bond, the oxime bond is more stable.
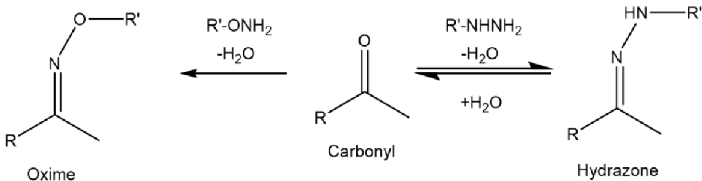
Figure 6: The Oxime and Hydrazone bonds.
A suitable enrichment strategy will allow analysis of complex mixtures giving us a better understanding of proteins and sites which are more susceptible to carbonylation. However, due to time contains on the project, a suitable tagging and enrichment method was not able to be established.
Conferences Attended
The 15th East Midlands Proteomics Workshop on the 16th of November 2016 at Nottingham Trent University.
The 4th Early Career Researchers Meeting in Analytical Biosciences on the 15th and 16th of March 2017 at the University of Warwick. Poster presented.
Syngenta Product Safety & Biology Research Collaborations Review Event on the 5th of September 2017 at Jealott’s Hill. Poster presented.
Syngenta Product Safety & Biology Research Collaborations Review Event on the 6th & 7th of September 2018 at Jealott’s Hill. Talk and poster presented.
The 39th BMSS annual meeting on the 10th-13th of September 2018 at Churchill College Cambridge. Poster presented.
The 17th East Midlands Proteomics Workshop on the 24th of October 2018 at Lincoln University. Poster Presented.
Advances in the Study of Lipid and Protein Oxidation: From Methods to Targets on the 13th-15th March 2019 at the University of Ghent Belgium. Talk and poster presented.
The 67th ASMS Conference on Mass Spectrometry and Allied Topics on the 2nd-6th of June 2019 in Atlanta, Georgia, USA. Poster presented.
Outreach
During open days I have run the Proteomics stand in the School of Life Sciences at Warwick. This involved demonstrating a simple time of flight mass spectrometer using ping-pong balls to both prospective students and their families. The families weren't necessarily from a scientific background, so the explanation had to be accessible to people at all levels, and not just those with a scientific background. The work also involved answering questions about proteomics, some at a higher level than others so an in-depth knowledge of the area was required.

The Proteomics stand at the Life Sciences open day at the University of Warwick.
MSc Project
Magnetic resonance project with Dr Jozef Lewandowski on "Development of Sample Preparation Methods to Study Antibody-Drug Conjugates". Antibody-drug conjugates can be used to directly target infected cells, with minimal side effects. There are currently two such conjugates on the market as an alternative to chemotherapy for cancer treatment. A new way of characterising them especially at an atomic level is needed before they can become more widely available. Sedimentation is a promising avenue and the project centres around the development of an ultracentrifuge tube which the sample can be sedimented in and then directly transferred to an NMR rotor using ultracentrifugation, thus reducing sample loss. Results were very successful and the tubes designed in the project were able to reach 700,000 g in the ultracentrifuge.
Other Academic Work
Undergraduate summer project on "Analysis of Ligand Binding to the Carbohydrate Recognition Domain of DC-SIGNR" in Chemical Biology with Dr Ann Dixon. DC-SIGNR is a c-type lectin which works as part of the immune system but also plays an important role in the spread of the HIV virus through the human body. The carbohydrate recognition domain of DC-SIGNR binds to gp120 on the surface of HIV, but it was unknown whether or not the binding was pH dependent. DC-SIGNR contains LL motifs involved in ligand internalisation but, does not contain YKSL motifs which its sister protein DC-SIGN contains. DC-SIGN using both these motifs is able to internalise its ligand and release it under low pH. The question was therefore raised as to whether the binding of DC-SIGNR is pH dependent or not. In order to answer this, a low pH titration of DC-SIGNR with mannose was done and results were studied using solution-state NMR. The findings are due to be published by Dr Ann Dixon.
Academic Societies
Member of the Royal Society of Chemistry since 2008
Member of the British Mass Spectrometry Society since 2016
Member of the British Society for Proteomic Research since 2019
Member of the American Society for Mass Spectrometry since 2019
Other Interests
Other than working for my PhD I like to spend time going to the gym with other members of MAS and friends from my lab. I watch and play a variety of different sports, read novels and watch anime in my spare time. Baking and other crafts help me to keep calm under the stress of my work. For the past year, I've been teaching myself Italian, but can understand more than I can speak. Since finishing my PhD I have started a postdoctoral position in the Thalassinos lab in the department of structural and molecular biology at UCL, using cross-linking mass spectrometry to analyse proteins associated with Huntington's disease.

Georgina Charlton
MAS CDT
Senate House
University of Warwick
Coventry
CV4 7AL
g dot charlton at warwick dot ac dot uk
